Database content
Database Content Overview
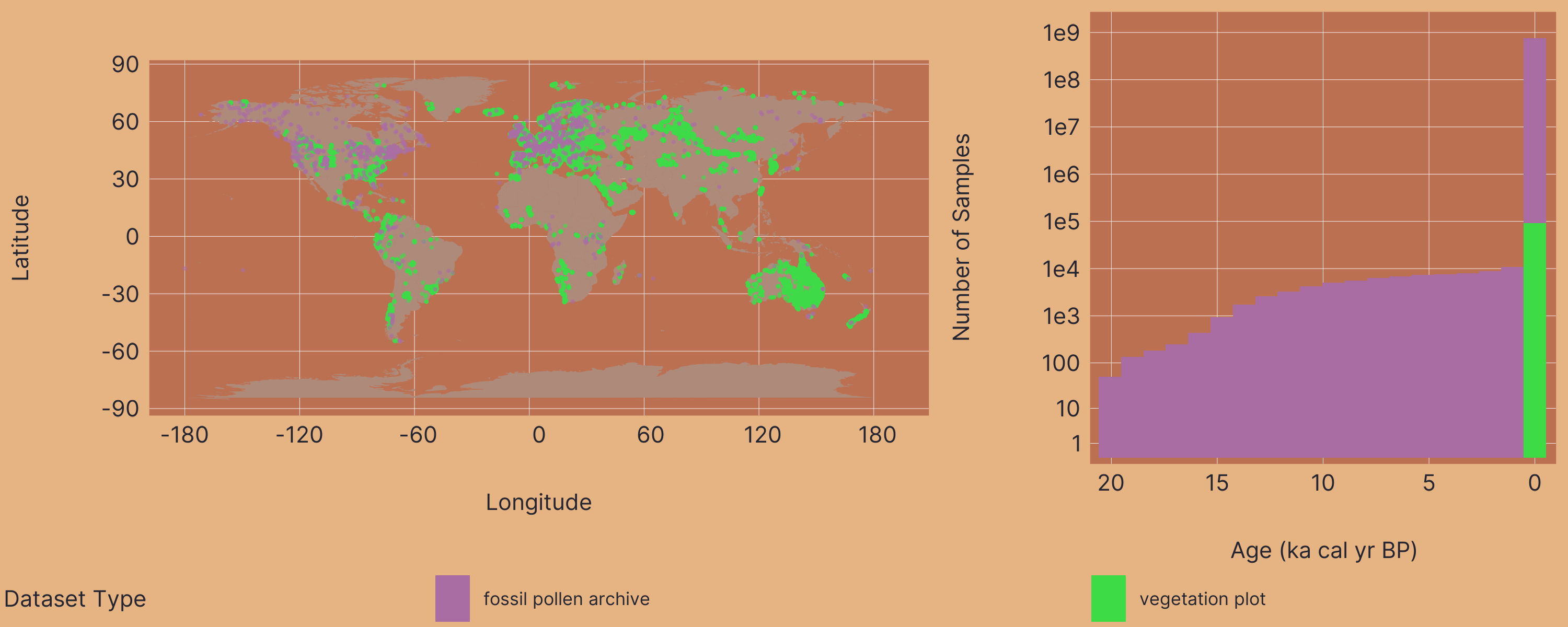
VegVault v1.0.0 is a comprehensive SQLite database (~110 GB) that integrates global vegetation data spanning both contemporary and paleovegetation records. This unified database combines fossil pollen records, contemporary vegetation plots, functional trait measurements, and associated abiotic environmental data to provide an unprecedented resource for vegetation ecology research.
The database consists of 31 interconnected tables with 87 fields (variables). For detailed technical information, see our Database Structure documentation.
Database Statistics
VegVault v1.0.0 contains:
| Datasets | > 480,000 |
| Samples | > 13,000,000 |
| Taxa | > 110,000 |
| Vegetation Traits | 6 |
| Trait Values | > 11,000,000 |
| Abiotic Variables | 8 |
| Geographic Coverage | Global |
| Temporal Coverage | 0-20,000 years BP |
Tip
Getting Started: New to VegVault? Check our Database Access guide for download instructions and our Usage Examples for practical applications.